Traits
Trait: Plague susceptibility and autoimmunity (ERAP2)
Dr Haran Sivapalan
/
November 28, 2022

What is antigen presentation?
Have you ever wondered how our immune system “knows” which cells to attack?
Bacteria, viruses, fungi, and other harmful microorganisms (collectively known as pathogens) have unique molecular markers that identify them as “foreign” or “non-self”. These markers, which include the components of the cell wall of bacteria, proteins that make up the flagellum (the swimming tail-like structure) of protozoa, or the protein coat of a virus, are called antigens.
By recognising and “remembering” specific antigens, our immune system can mount an appropriate response to particular pathogens and quickly target them the next time they enter the body. This is the principle behind adaptive immunity, and underpins why vaccinations help protect us against disease.

One type of white blood cell (lymphocytes), known as B cells, has specialised antigen receptors, called B-cell receptors (BCRs) that bind to whole antigens on bacteria etc.. When this happens, B cells become activated, multiply, and start making antibodies that are targeted to the specific antigen and help neutralise the invading organism. This is illustrated in the image above.
We also have another type of white blood cell, known as T cells. Rather than producing antibodies, T cells help destroy infected cells and help coordinate a wider immune response to a pathogen.

There are two classes of T cell:
- Cytotoxic T cells (also known as CD8+ T cells) - which destroy cells infected with pathogens.
- T helper cells (also known as CD4+ T cells) - which secrete signaling molecules (cytokines) that activate other immune cells and help coordinate a wider immune response. T helper cells are so-called because they help B cells produce antibodies and help cytotoxic T cells to kill infected cells.
T cells also have antigen receptors, called T cell receptors (TCRs), that can recognise the specific antigens on bacteria etc. Unlike those of B cells, the antigen receptors on T cells cannot bind to whole antigens.
Instead, antigens (say a protein on the surface of a bacteria) must first be broken down into smaller bits, before being presented to T cells so that they can orchestrate a targeted immune response.
This process of chopping up, processing, and presenting antigens to T cells is known as antigen presentation.
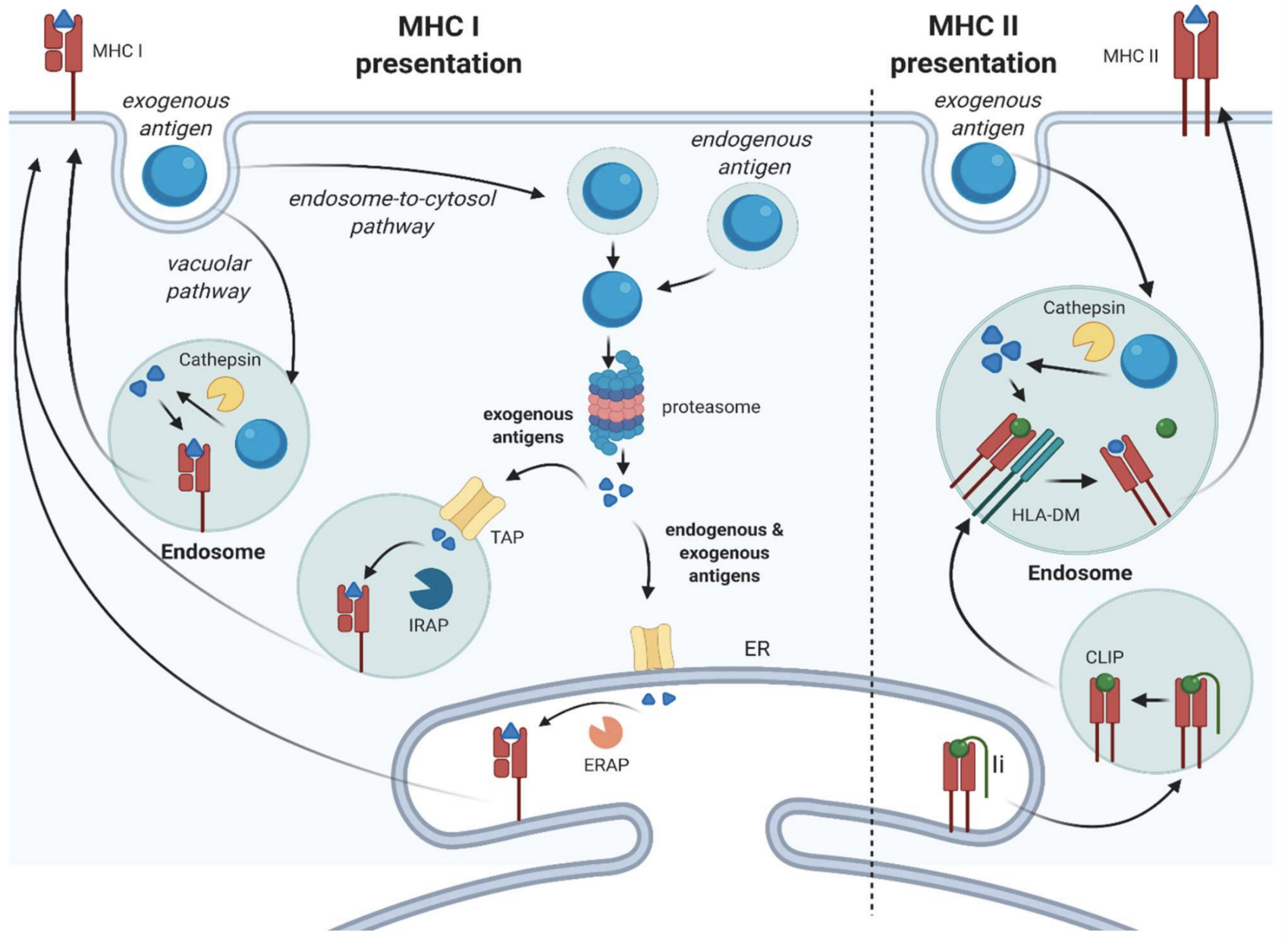
- The basics of antigen presentation
When our own cells become infected by, say, a bacterium, they chop up the bacterial antigen into smaller pieces (called antigen peptides) using specialised enzymes. These smaller antigen peptides, which can still uniquely identify the bacterium, are then combined with a protein from our own cells called MHC (Major Histocompatibility Complex).
The bacterial antigen peptide and MHC protein together are then displayed on the surface of the infected cell, akin to a sort of immunological barcode. T cells can then “scan” this barcode using their T-cell receptor. This allows T cells to recognise other infected cells, so that they can either be directly or indirectly destroyed / lysed (by cytotoxic T cells and T helper cells, respectively).

Source: Introduction to the Immune Response. Primer to the Immune Response. 2014:3–20. doi: 10.1016/B978-0-12-385245-8.00001-7. Epub 2014 Oct 10. PMCID: PMC7184560.
There are two classes of MHC protein that our cells use for presenting antigens, which depend on how a cell is infected and whether the antigen (or, more accurately, antigen peptide) is being presented to a cytotoxic T cell or a T helper cell.
- MHC Class I is used for bacteria, viruses, and other pathogens that have invaded cells (intracellular pathogens). It is used to present antigen peptides from these intracellular pathogens to cytotoxic T cells. Virtually all of our own cells can produce MHC Class I, so any cell that has been infected by a pathogen can be targeted for destruction by cytotoxic T cells. Immune cells such as macrophages can also engulf infected cells and present antigens using MHC Class I to cytotoxic T cells.
- MHC Class II is used for intracellular as well as extracellular pathogens. It is used to present antigen peptides to T helper cells. Extracellular pathogens may need to be engulfed by specialised immune cells (such as dendritic cells (DCs) or macrophages), before their antigen can be presented to T helper cells. Only certain kinds of immune cells (collectively known as antigen-presenting cells) produce MHC Class II. MHC Class I
If you're interested in reading about the various cells of the immune system, antigen presentation, and how we acquire immunity in more depth, this primer in the Elsevier Public Health Emergency Collection is a great article.
What is ERAP2?
ERAP2 (endoplasmic reticulum aminopeptidase 2) is an enzyme involved in our immune response to infection.
More specifically, ERAP2 is involved in presenting antigens from pathogens that have infected cells to T cells, facilitating the immune destruction of these infected cells.
Source: Harryvan, T. J., de Lange, S., Hawinkels, L. J. A. C., & Verdegaal, E. M. E. (2021). The ABCs of Antigen Presentation by Stromal Non-Professional Antigen-Presenting Cells. International Journal of Molecular Sciences, 23(1), 137. MDPI AG. Retrieved from http://dx.doi.org/10.3390/ijms23010137
As explained in the previous section, antigens from bacteria, viruses, and other pathogens need to be chopped up into smaller antigen peptides and then bound to MHC proteins before they can be presented to T cells.
The ERAP2 enzyme in particular is involved in the final trimming of antigen peptides before they are loaded onto MHC Class I protein.
The resultant complex of MHC Class I protein and antigen peptide can then be presented to cytotoxic T cells. This allows cytotoxic T cells to recognise and destroy other cells infected by the same pathogen, as such infected cells will also express the same antigen peptide.
The ERAP2 enzyme is encoded by your ERAP2 gene.
How do ERAP2 gene variants affect immune response to infection?
A SNP (single nucleotide polymorphism) in the ERAP2 gene, designated rs2549794, creates two different ERAP2 gene variants or alleles:
- the ‘C’ variant
- the ‘T’ variant.
Studies have shown that the ‘C’ variant is linked to 5 times higher ERAP2 expression compared to the ‘T’ variant. For example, in one study, immune cells from people with either no, one, or two copies of the ‘C’ variant (TT, CT, CC genotypes, respectively) were analysed to assess for differences in expression of the ERAP2 gene. (Gene expression is essentially the process by which the instructions in a gene are read and used to make the encoded protein. We can roughly say that higher ERAP2 gene expression results in greater production of the ERAP2 enzyme by various cells).
The researchers looked at both baseline levels of gene expression (unstimulated / non-infected condition), as well as gene expression when the cells were incubated with Yersinia pestis - the bacterium that causes plague.
As illustrated in the graphs below, the immune cells of people with one or two copies of the C variant (CT and CC genotypes) showed significantly higher ERAP2 expression compared to those from people with the TT genotype. This was true both when the immune cells were unstimulated/non-infected and in the context of Y.pestis infection.


Source: Klunk, J., Vilgalys, T. P., Demeure, C. E., Cheng, X., Shiratori, M., Madej, J., ... & Barreiro, L. B. (2022). Evolution of immune genes is associated with the Black Death. Nature, 1-8.
The same study also incubated white blood cells (macrophages) with Yersinia pestis bacteria to see how well the white blood cells killed and prevented the bacteria from replicating.
As shown in the boxplots below, white blood cells taken from those with the CC genotype killed the greatest percentage of bacteria, followed by the CT genotype and then the TT genotype.

Source: Klunk, J., Vilgalys, T. P., Demeure, C. E., Cheng, X., Shiratori, M., Madej, J., ... & Barreiro, L. B. (2022). Evolution of immune genes is associated with the Black Death. Nature, 1-8.
These results suggest overall that, by enhancing expression of the ERAP2 gene, the ‘C’ variant (rs2549794) facilitates a more effective immune response to various pathogens, including Yersinia pestis bacteria.
More specifically, it is thought that higher ERAP2 expression linked to the ‘C’ variant allows more effective presentation of bacterial antigens to cytotoxic T cells (CD8+ cells). This may enable cytotoxic T cells to more easily destroy infected cells, helping to clear bacterial infections more quickly.
What is plague?
Plague is an infectious disease caused by the Yersinia pestis bacterium, which is typically found in small mammals (particularly rodents) and their fleas.
Humans become infected with the bacteria when bitten by an infected flea. The bacteria can also be spread through unprotected contact with infectious bodily fluids or contaminated materials (e.g. from handling infected animal tissue), or breathing in respiratory droplets of an infected person (with pneumonic plague).

Image of plague buboes in the groin. Source:Wikimedia Commons
The general symptoms of plague include fever, headache, chills, and weakness, while more specific symptoms differ according to the type of plague and route of infection. On this note, there are three main forms of plague:
- Bubonic plague - the most common form of plague, which is caused by the bite of an infected flea. Bacteria from the flea bite travel to nearby lymph nodes and cause them to become swollen, painful, and inflamed. These swollen lymph nodes are known as “buboes.”
- Pneumonic plague - this form of plague affects the lungs, and is caused by inhaling the respiratory droplets of other people with pneumonic plague. Bubonic plague that is left untreated and spreads to the lungs can also cause pneumonic plague.
- Septicaemic plague - this is when plague infection spreads in the bloodstream. It can cause bleeding and lead to septic shock.
Historically, various plague pandemics have killed millions across the world. Perhaps the most infamous pandemic was the Black Death: the pandemic that affected Europe, the Middle East, and North Africa between 1346 and 1350 AD, reducing the population by 30 - 50%.
Plague still infects people today, occurring in several countries in Africa, North America, South America, and Asia. Worldwide, there were 3,248 cases of plague reported between 2010 and 2015. Since the 1990s, most plague cases have occurred in Africa, with Madagascar having the highest incidence of cases.
Fortunately, plague can be treated and effectively cured with common antibiotics. Pneumonic plague that is left untreated, however, can rapidly become fatal.
For more information on plague, the CDC (Centers for Disease Control and Prevention) and WHO (World Health Organisation) pages are good resources.
How did ERAP2 gene variants affect susceptibility to the Black Death?
A recent study suggests that the ‘C’ variant (rs2549794) of the ERAP2 gene affords greater protection against Yersinia pestis infection and plague, with ‘C’ variant carriers historically having been more likely to survive the Black Death plague pandemic (1346 - 1350 AD).
The study dated and examined DNA samples from people buried at various cemeteries in London and across Denmark. These individuals had either died before (850 - 1250 AD), during (1348-1349), or after (1350 -1800AD) the Black Death.
According to the evolutionary theory of natural selection, if a gene variant increases survival, we would expect this variant to be selected for and therefore increase in frequency in the population over generations. On this basis, if a gene variant promotes survival during a plague pandemic, we would expect this beneficial variant to be more commonly seen in individuals who died after the Black Death compared to those who died before and during the Black Death.
Conversely, if a particular gene variant made someone more susceptible to plague, we would expect the frequency of this variant in populations to be significantly lower after the Black Death. This is because people carrying this disadvantageous variant would have been less likely to survive the Black Death and pass on their genes to future generations.

Source: Source: Klunk, J., Vilgalys, T. P., Demeure, C. E., Cheng, X., Shiratori, M., Madej, J., ... & Barreiro, L. B. (2022). Evolution of immune genes is associated with the Black Death. Nature, 1-8.
As illustrated in the diagram above, the frequency of the ‘C’ variant of the ERAP2 gene was found to be significantly higher in people who died after the Black Death compared to those who died before and during the Black Death. (The blue markers are for DNA samples from Denmark; the red markers are for samples from London burial sites).
This strongly suggests that the ‘C’ variant was protective against Yersinia pestis infection and enhanced survival during the Black Death plague pandemic. In fact, the researchers calculated that people with two copies of the ‘C’ variant (i.e. those with the CC genotype) were roughly 40% more likely to survive the Black Death compared to those with no copies (i.e. those with the TT genotype).
The survival advantage of the ‘C’ variant is likely due to its effects on ERAP2 expression (as explained previously). ‘C’ variant carriers with higher ERAP2 expression may have been better able to present antigens from the Yersinia pestis bacterium to cytotoxic T cells. This would support a stronger immune response and more effective destruction of cells infected with the bacteria.
How do ERAP2 gene variants affect susceptibility to other infections?
A recent preprint study associated the ‘T’ variant (rs2549794) of the ERAP2 gene with greater susceptibility to respiratory infections such as pneumonia.
The study collated the data from three large study cohorts (UK Biobank, FinnGen, and GenoMICC) and looked at the ERAP2 genotypes of people (cases) admitted to hospital for pneumonia, severe COVID-19, and sepsis. The frequency of ERAP2 genotypes was then compared to those of healthy controls.
If a particular ERAP2 gene variant increases susceptibility to respiratory infection, we would expect this variant to be more frequently seen in cases compared to controls. For carriers of this risk variant, the odds of being a case rather than a healthy control would be greater relative to non-carriers.

Source: Hamilton, F. W., Mentzer, A. J., Parks, T., Baillie, J. K., Smith, G. D., Ghazal, P., & Timpson, N. J. (2022). Variation in ERAP2 has opposing effects on severe respiratory infection and autoimmune disease. medRxiv.
As can be seen in table above, the ‘T’ variant was, relative to the ‘C’ variant, associated with a 2.5% higher odds of pneumonia, a 7.8% higher odds of critical care admission for pnemonia, and a 2.7% higher odds of severe COVID-19. The ‘T’ variant did not however appear to increase the risk of sepsis.
The study authors suggested that relatively lower ERAP2 expression caused by the ‘T’ variant leads to less effective presentation of bacterial and viral antigens to cytotoxic T cells, and a weaker inflammatory cytokine response. This would contribute to a less effective immune response to certain pathogens, making ‘T’ variant carriers more susceptible to respiratory infection.
How do ERAP2 gene variants affect susceptibility to Crohn’s disease and other autoimmune conditions?
Crohn’s disease is an inflammatory bowel disease involving chronic inflammation of the digestive system.
The disease is thought to be an autoimmune condition, whereby the immune system attacks the body’s own cells. Normally, cells of the immune system recognise the antigens on our own cells (self-antigens) and those of harmless organisms (e.g. beneficial gut bacteria), and accordingly do not mount an immune response against these cells. This capacity to not attack our own cells is known as immunological tolerance.
In autommine conditions such as Crohn’s disease, however, there is a loss of immunological tolerance, which causes the immune system to attack cells expressing self-antigens. Cytotoxic T cells primed against these self-antigens will try to destroy these cells, while T-helper cells will orchestrate an inflammatory response, and B cells may produce antibodies targeted to the self-antigens (known as autoantibodies).

In Crohn’s disease specifically, the immune system is thought to attack self-antigens in the intestinal lining. T cells are also directed against harmless gut bacteria. Both of these autoimmune processes lead to inflammation and tissue damage of the bowel wall.
- ERAP2 variants and Crohn’s disease
Some studies suggest that the ‘C’ variant (rs2549794) of the ERAP2 gene, which affords greater protection from plague and other respiratory infections, is linked to a greater risk of Crohn’s disease.
A 2010 meta-analysis, which compared the ERAP2 genotypes of 6,333 people with Crohn’s disease to 15,056 healthy controls, found that the ‘C’ variant was associated with a 5% higher odds of having Crohn’s disease..
Complementing this finding, a recent preprint study found that the ‘T’ variant was linked to a 14.4% lower odds of Crohn’s disease. The same study also associasted the ‘T’ variant with a 5.4% lower risk of Type I diabetes - another autoimmune disease.




.png)
