Traits
Trait: DNA repair and longevity (TP53)
Dr Haran Sivapalan
/
January 3, 2022

What are tumour suppressor genes / proteins?
For us to grow, develop, and maintain normal tissue function, our cells need to divide and replicate.
Take our skin as an example. If you’ve ever cleaned a dusty room, you may not be surprised to learn that we shed about 30,000 to 40,000 skin cells every minute (household dust being famously claimed to be largely composed of dead skin cells). As a result of such high turnover, our skin cells need to divide regularly to replenish the lost cells and maintain normal skin function.
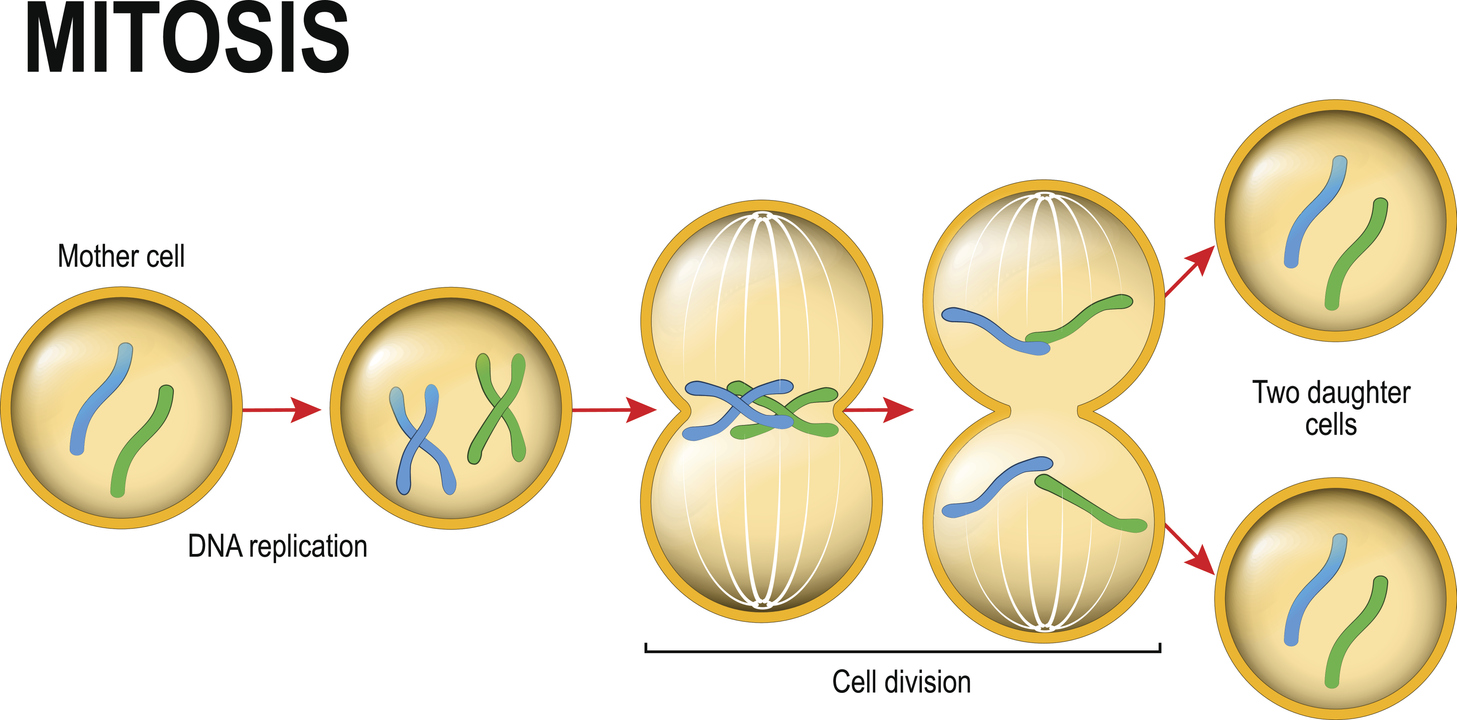
Of the estimated 37 trillion (i.e. 37,000,000,000,000) cells in our body, about 2 trillion divide every day. This process of cell division (known as mitosis), however, needs to be regulated. If cells were allowed to divide uncontrollably, we would end up with excess tissue that would soon compromise our health. For example, without checks on cell division, it’s thought that an 80-year-old person would accumulate 2 tonnes of bone marrow and lymph nodes, and develop intestines measuring 16-km long!
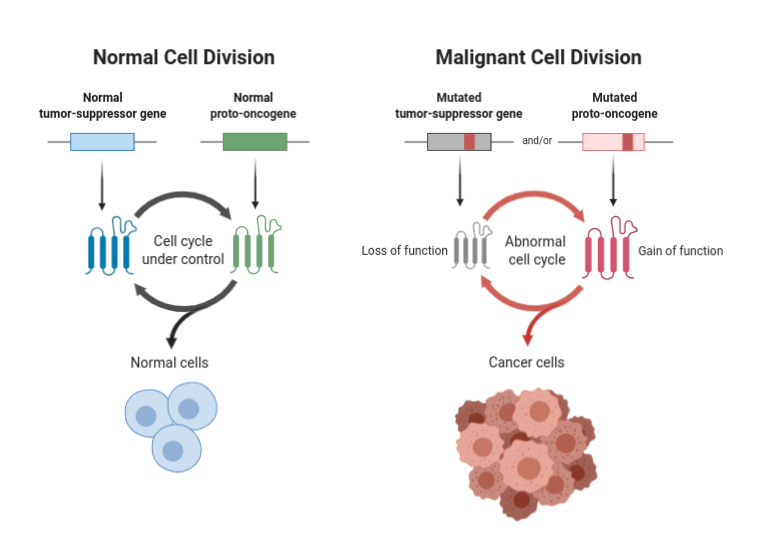
More gravely, if a group of cells were to divide uncontrollably, they could spread and invade other organs, impairing their function. This is essentially what happens in a malignant tumour, where an abnormal mass of cells spreads to other parts of the body.
Another reason to regulate cell division is to stop damaged cells from multiplying and perpetuating future generations of damaged cells. If a cell sustains DNA damage or inherits mistakes in its genetic code, these DNA errors (if left unrepaired) will be passed on to its daughter cells when it divides. If left unchecked, tissues and organs would then be increasingly comprised of damaged cells and may cease to function effectively.
We’ve now explained why we need to regulate cell division, but how exactly do we go about this?
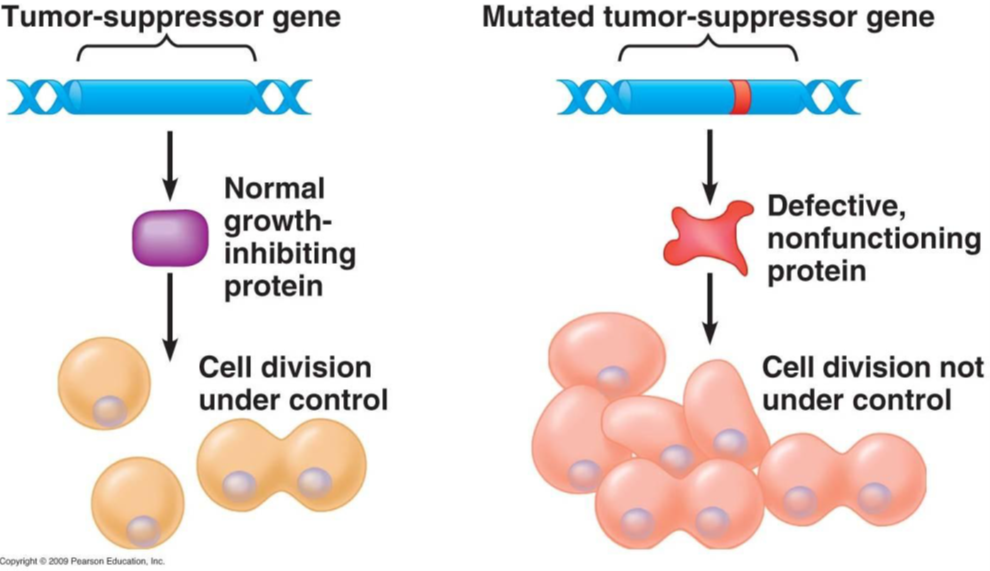
Enter tumour suppressor genes. These are specialised genes that, when activated, act like a brake to slow down or halt cell division. Tumour suppressor genes encode proteins, called tumour suppressor proteins, that prevent cells from dividing and replicating. Such proteins are said to be acting as “gatekeepers” of cell division.
Tumour suppressor proteins can also repair damaged DNA (known as “caretaking”) or cause damaged cells to “commit cell suicide” via a process of programmed cell death called “apoptosis”. We’ll talk more about apoptosis in the next section.
If tumour suppressor genes are inactivated or mutated, however, they may no longer be able to control cell division. Consequently, cells in which tumour suppressor genes are inactivated can divide uncontrollably, forming abnormal masses of cells: tumours.
Source:BioRender (2021). Tumor Suppressor Genes and Proto-oncogenes. Retrieved from https://app.biorender.com/biorender-templates/t-5ed6b0f2cc82d300ae329495-tumor-suppressor-genes-and-proto-oncogenes
In fact, this is from where the term “tumour suppressor gene” emanated. Studies of retinoblastoma, a rare childhood eye tumour, found that the tumour cells had two inactivated copies of a gene called Rb. Further laboratory experiments that inserted normal Rb genes into retinoblastoma cells found that it prevented them from dividing uncontrollably. The normal, active variant of the Rb gene was therefore suppressing the formation of retinoblastoma tumours.
Since then, several other tumour suppressor genes have been identified, including: APC, BRCA1, BRCA2, and the subject of this trait, p53. These genes all play a role in regulating cell division, preventing uncontrolled cell growth, and repairing damaged DNA.
Rare mutations within these genes can give rise to or increase susceptibility to various cancers. For example, mutations within the APC gene can give rise to a condition known as Familial Adenomatous Polyposis (FAP), characterised by hundreds to thousands of small growths (polyps) in the colon. If left untreated, these can develop into colorectal cancer.
As a general rule, FitnessGenes do not report on rare gene mutations that are strongly linked to cancer. This is because we believe these types of DNA results require appropriate genetic counselling and discussion with trained health professionals.
In this respect, we will only look at common SNPs in the P53 gene that do not have robust links to cancer risk.
KEY POINTS
- Cells have protective mechanisms to ensure damaged and mutated cells do not replicate and pass on mistakes.
- Tumour suppressor proteins are specialised proteins that suppress cell division and protect against uncontrolled cell growth.
- Tumour suppressor proteins are coded for by tumour suppressor genes.
- Mutations in tumour suppressor genes can lead to uncontrolled cell growth and the development of cancers.
- FitnessGenes do not analyse mutations with a strong link to cancer as we believe such information should be provided only with appropriate genetic counselling.
What is p53?
One of the best studied tumour suppressor proteins is p53.
It has been widely dubbed the “guardian of the genome” as it helps to repair damaged DNA and acts as a guardian or gatekeeper to stop cells with damaged and mutated DNA from dividing.
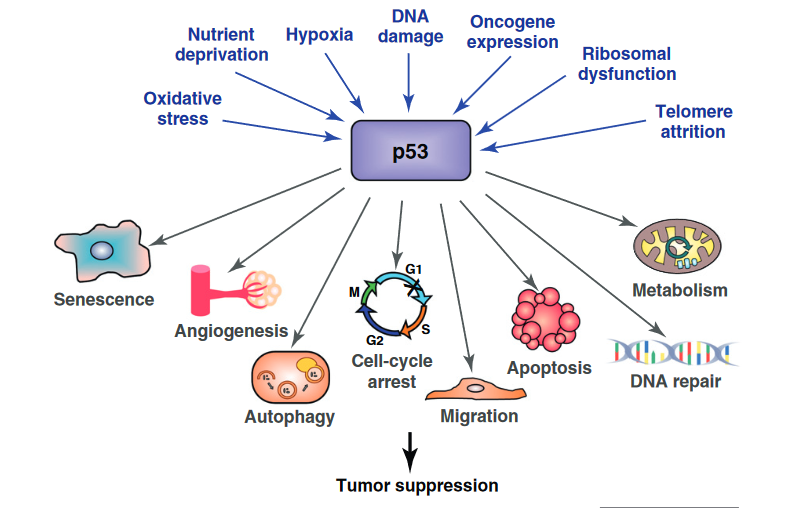
p53 is activated in response to DNA damage and other types of cellular stress, such as low oxygen (hypoxia), nutrient deprivation, and oxidative stress.
In response to being activated, p53 switches on genes that fixed damaged DNA, slow or arrest cell division, and initiate other changes that prevent uncontrolled cell growth and the formation of tumours. Collectively these protective responses to DNA damage are known, aptly, as our DNA damage response.
Source: Bieging, K. T., & Attardi, L. D. (2012). Deconstructing p53 transcriptional networks in tumor suppression. Trends in cell biology, 22(2), 97-106.
In particular, p53 activates three different strands of our DNA damage response.
- p53 activates DNA repair pathways to fix regions of damaged DNA.
- p53 “arrests” damaged cells at checkpoints in their cell division cycle to prevent them from dividing (cell cycle arrest).
- p53 causes heavily damaged cells to eliminate themselves via programmed cell death (apoptosis).
By repairing damaged DNA, preventing damaged cells from dividing, and destroying irreparably damaged cells by apoptosis, p53 is thought to play a major role in preventing uncontrolled cell growth and the development of cancer. Let’s look at these three processes in further detail.
DNA repair
As explained in the ATM and DNA damage trait article, DNA molecules in our cells can sustain various kinds of damage, such as breaks to one or both strands (single and double-strand breaks), abnormal links between strands, incorrect pairing of bases (bases being the “letters” that make up the DNA sequence), or the insertion or deletion of bases.
If left unrepaired, these unwanted changes to the structure and sequence of DNA molecules could lead to the production of faulty proteins, interfere with important cell processes, and ultimately impair cell and tissue function.
Luckily, as outlined in the ATM and DNA damage trait, our cells have various repair mechanisms that can fix damaged DNA. For example, we have various enzymes that can make a nick in a DNA molecule, excise any damaged bases, and then reseal the DNA molecule, much in the same way a surgeon may open and then debride an infected wound of rotten tissue before stitching it back up.
In response to DNA damage, levels of the p53 protein quickly rise. p53 then activates the various enzymes and repair pathways responsible for fixing damaged DNA.
Cell cycle arrest
- The cell cycle
It’s an obvious oversimplification that is not applicable to all, but the life cycle of a human can be said to involve being born as a baby, spending several years growing up and developing, finding a partner, and then eventually birthing a new baby to repeat the cycle.
By a similar token, the life cycle of an individual cell involves being formed by division from its mother cell, growing in size, replicating its DNA, before dividing (by mitosis) to form two new daughter cells of its own.
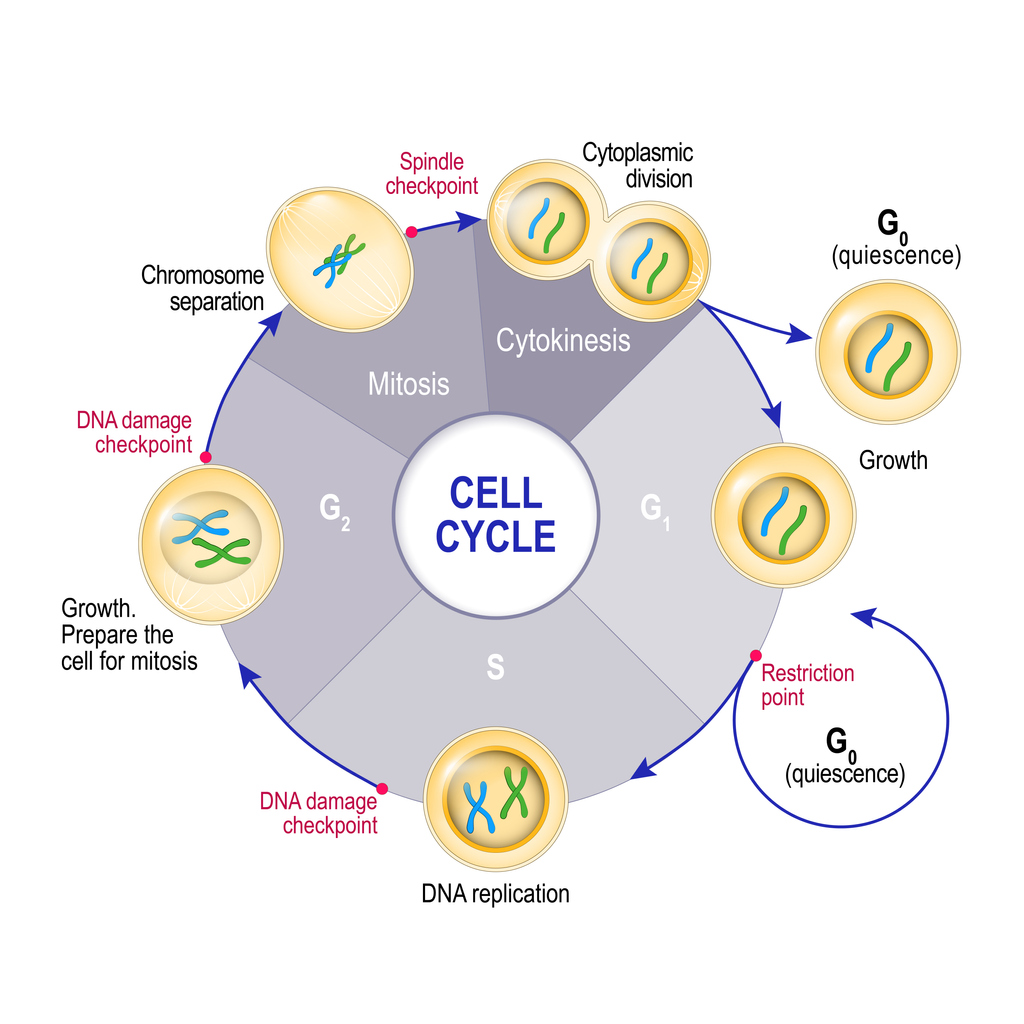
This cycle of growth and division is known as the cell cycle. The cell cycle can be split into 4 distinct phases:
- G1 (first gap phase) – a newly formed cell grows larger, accumulates nutrients, and develops its organelles. At the end of this phase, the cell can “decide” whether it wants to exit the cycle and go into a resting or ‘quiescent’ state (G0), or continue into the next phase of the cell cycle (S) and prepare to divide into new daughter cells.
- S (DNA synthesis phase) – the cell replicates its DNA (which is packaged into chromosomes) to create genetic code for two new daughter cells.
- G2 (second gap phase) – the cell grows further and develops organelles in preparation to divide by mitosis into two daughter cells.
- M (mitotic phase) – the duplicated DNA / chromosomes are pulled apart (chromosome separation) and the cytoplasm of the cell splits into two (cytokinesis), forming two new daughter cells. The cycle can then start again.
Cell cycle checkpoints
As stressed earlier, if a damaged or mutated cell were allowed to progress through the cell cycle without any checks, any mistakes in its DNA would be passed on to its new daughter cells as it divided. As these daughter cells then restart their own cell cycles, these mistakes could quickly proliferate, leading to the accumulation of abnormal cells.
To prevent damaged cells from dividing, the cell cycle has various in-built checkpoints to ensure everything is going to plan. You can think of these checkpoints as similar to the series of security checks when flying to another country (e.g. check-in, carry-on baggage screening, boarding gate, border/passport control etc).
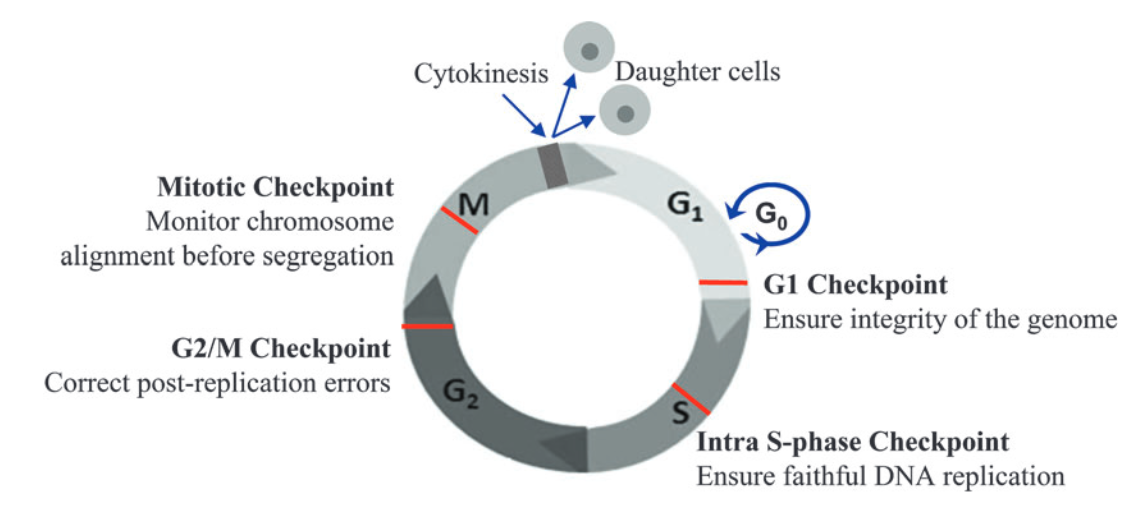
Source: Ma W. (2014) Cell Cycle Checkpoint. In: Schwab M. (eds) Encyclopedia of Cancer. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-662-46875-3_996
There are 4 key checkpoints that a cell passes through in the cell cycle:
- G1 checkpoint – this checkpoint occurs at the end of the G1 phase, at the G1/S transition. It checks that the cell is developed and large enough to divide, has enough stored nutrients for cell division, and, importantly, whether there is any DNA damage. Once a cell is allowed to pass the G1 checkpoint, it is essentially committed to progress through the rest of the cell cycle and divide. The G1 checkpoint is therefore crucial and is known as a restriction point.
- Intra-S checkpoint – this checkpoint occurs during the S phase to check for DNA damage during replication.
- G2/M checkpoint – this checkpoint occurs at the end of the G2 phase, just before the M phase. It checks whether DNA has been correctly replicated in the S phase and whether there is any DNA damage. Any mistakes in DNA molecules at this stage will end up in the two new daughter cells formed during mitosis, making the G2/M another key checkpoint.
- Mitotic (spindle) checkpoint – this checkpoint ensures chromosomes are correctly aligned to be pulled apart into two new daughter cells during mitosis.
So, what happens if DNA damage is detected at the various checkpoints of the cell cycle?
The two main checkpoints controlled by p53 are the G1 and G2/M checkpoints. If DNA damage is sensed at either of these checkpoints, p53 will stop the cell from progressing through the rest of the cell cycle. This is known as cell cycle arrest.
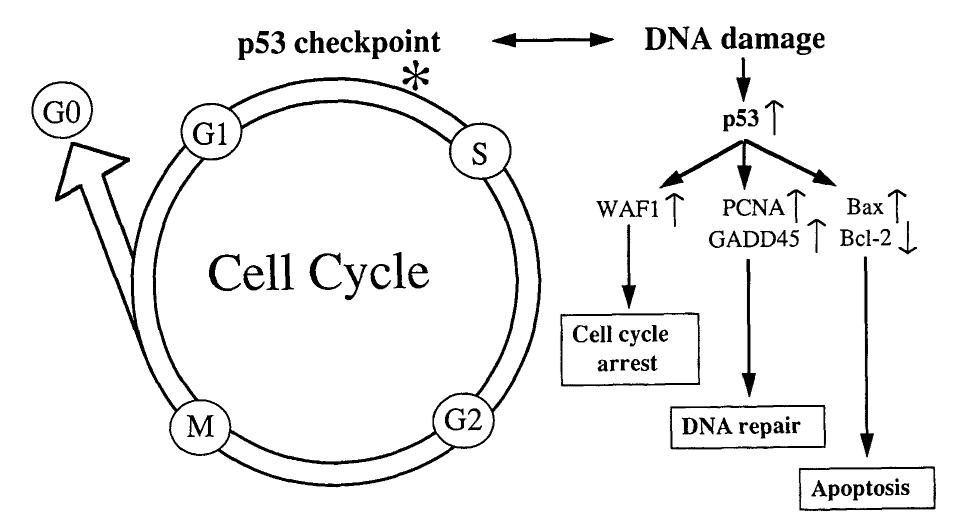
Source: Baas, I. O., Offerhaus, G. J. A., & Hruban, R. H. (1997). Interactions between cytomegalovirus and the p53 tumor suppressor gene in atherosclerotic vascular disease. Clinical Microbiology and Infection, 3(4), 393-395.
While halted at these checkpoints, p53 can then initiate DNA repair mechanisms (explained above) to fix any damaged regions of DNA. If the damaged DNA is successfully repaired, the cell may then be allowed to progress to the next phase of the cell cycle.
At the G1 checkpoint in particular, cells may be made to exit the cycle and enter the G0 quiescent phase. In this G0 phase, the cell does not divide but remains in a “resting state” where it remains metabolically active and functions normally.
Some cells enter this state only temporarily and can re-enter the cell cycle when conditions are appropriate – this is known as quiescence. By contrast, damaged cells may be made to enter the G0 phase irreversibly, being permanently unable to grow and divide. This is known as senescence and is also regulated by p53. By forcing damaged cells to become senescent and unable to divide, p53 helps to prevent uncontrolled growth of abnormal cells.
In some cases, a cell may sustain may a heavy degree of DNA damage such that it cannot be repaired. In these cases, rather than being temporarily halted at a cell cycle checkpoint, p53 instructs the cell to destroy itself. This process of programmed cell death is known as apoptosis.
Apoptosis
Apoptosis, colloquially referred to as “cell suicide”, is a process of programmed cell death.
During apoptosis, cells contract and develop “blebs” on their surface, while the nucleus containing genetic material condenses. The cell then splits into small fragments known as apoptotic bodies, which are then engulfed by white blood cells (a process called phagocytosis).
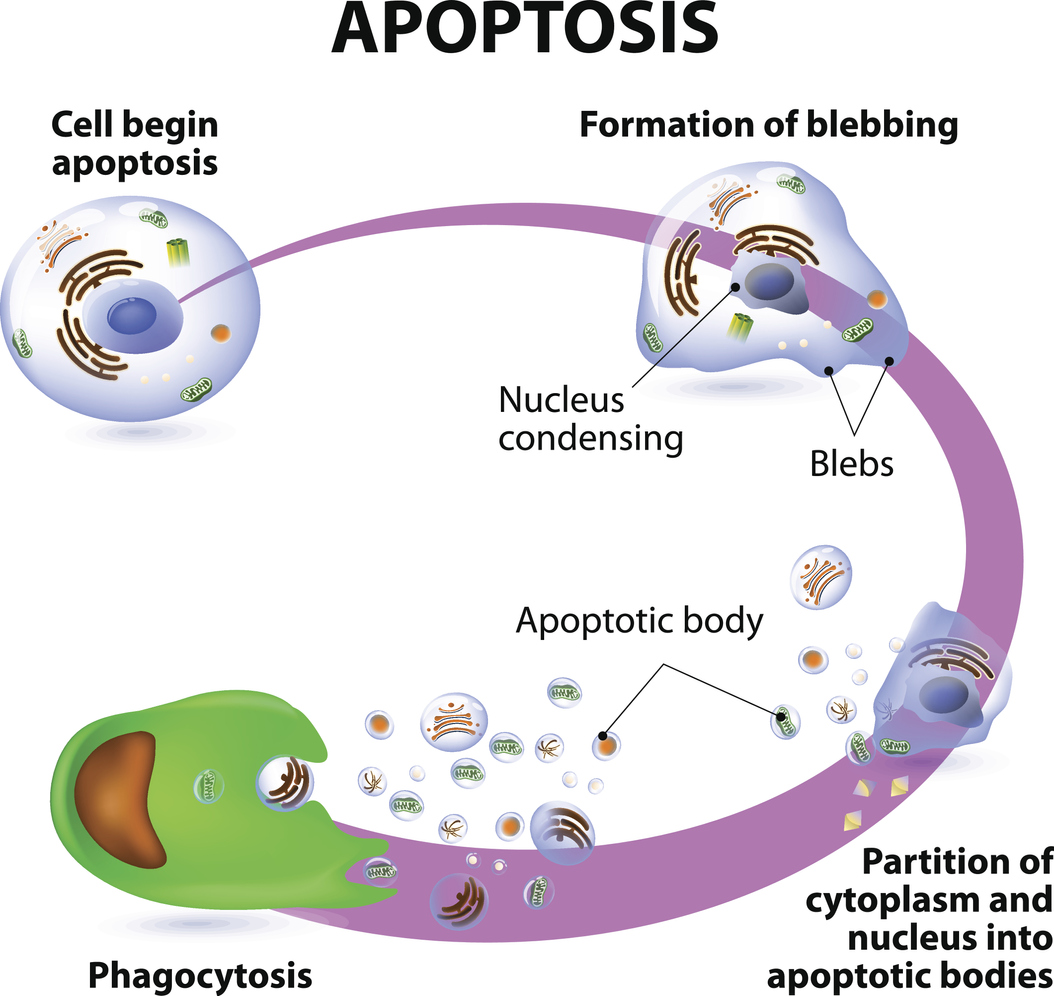
While apoptosis may sound like a pathological event, it is actually a normal, controlled process that is often used to eliminate cells that are no longer needed. Cells in developing foetuses undergo apoptosis during the modelling of tissues and organs, for example.
Apoptosis is also used to eliminate cells that may pose a threat if allowed to divide. For example, cells infected with a virus are often targeted for apoptosis to prevent the spread of infection to other sites in the body.
Cells with heavy DNA damage are also destroyed by apoptosis. After arresting damaged cells at the G1 and G2/M checkpoints, p53 can target any cells with irreparable DNA damage for apoptosis. If these cells were not eliminated, they may progress through the cell cycle, divide, and pass on DNA damage to future cells, causing the accumulation of abnormal cells.
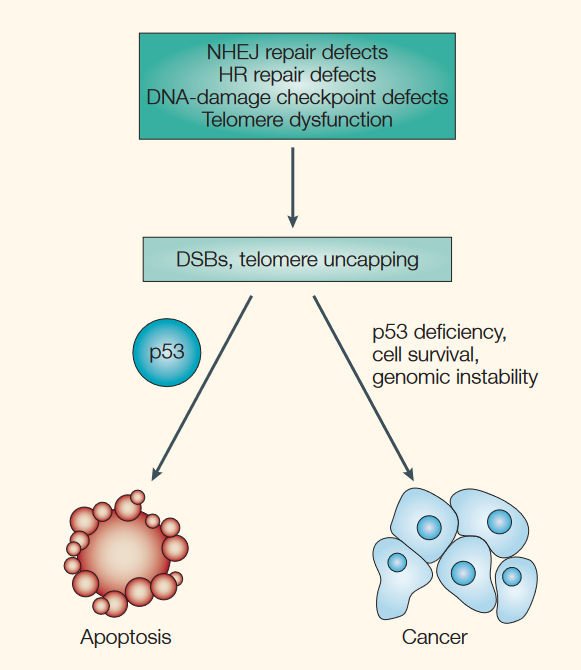
Source: Brown, J. M., & Attardi, L. D. (2005). The role of apoptosis in cancer development and treatment response. Nature reviews cancer, 5(3), 231-237.
On this note, tumour cells are often observed to have defective p53 function, which allows them to evade apoptosis and divide uncontrollably. By stimulating apoptosis, p53 helps to prevent the development of tumours. Similarly, chemotherapy and radiation used in the treatment of cancer work partly by stimulating cancer cells to undergo apoptosis.
KEY POINTS
- p53 is one of the most widely studied tumour suppressor proteins in cells.
- p53 acts to suppress division of damaged cells and prevent uncontrolled cell growth.
- p53 is activated in response to DNA damage and other forms of cell damage e.g. low oxygen, nutrient depletion.
- Once activated, p53 stimulates DNA repair pathways to fix damaged DNA.
- p53 also "arrests" or halts damaged cells at checkpoints in their cell division cycle, preventing them from replicating.
- Heavily damaged cells that cannot be repaired are stimulated by p53 to 'commit suicide' through apoptosis - programmed cell death.
- Cells that escape cell cycle arrest and apoptosis may divide uncontrollably and become cancerous.
What is the TP53 gene?
p53 is coded for by our TP53 gene.
To adopt biological parlance, if p53 is a ‘tumour suppressor protein’, then TP53 is the corresponding tumour suppressor gene. The TP53 gene has been widely studied following observations that several different types of cancer cells are shown to carry TP53 mutations.
So-called somatic mutations of the TP53 gene, which are non-inherited and are formed after conception, are one of the most common genetic alterations in cancerous cells. For example, healthy cells exposed to UV radiation or carcinogenic chemicals may sustain TP53 mutations that cause them to become abnormal and divide uncontrollably.
This is likely due to such mutations impairing the function of p53, which means the mutated cells are not repaired, not arrested at cell cycle checkpoints (and thus prevented from dividing), and not targeted for self-destruction by apoptosis.
It is important to stress again that your DNA repair and longevity trait does not look at somatic or rare inherited TP53 mutations that are strongly linked to defective p53 function and cancer.
By contrast, we look at common variants of the TP53 gene that may slightly alter p53 activity but do not cause defective p53 function and are not robustly linked to cancer.
TP53 variants
A common SNP (Single Nucleotide Polymorphism) in the TP53 gene, designated rs1042522 or Arg72Pro, causes a G-->C change in the DNA sequence. This creates two different TP53 gene variants or ‘alleles’: ‘G’ and ‘C’.
The ‘G’ allele codes for the amino acid arginine at position 72 in the p53 protein and therefore is also called the Arg (Arg72) allele.
By contrast, the ‘C’ allele codes for proline at the same position and may therefore be referred to as the Pro (Pro72) allele.
Studies suggest that the ‘G’ (Arg) and ‘C’ (Pro) alleles can alter p53 activity, changing the way in which p53 activates DNA repair, cell cycle arrest, and apoptosis in response to DNA damage. This, in turn, may influence lifespan and longevity.
KEY POINTS
- The TP53 gene encodes the p53 tumour suppressor protein.
- Mutations in the TP53 gene that are inflicted after conception and not inherited (somatic mutations) are observed in cancer cells.
- Your DNA repair and longevity trait looks at common, inherited variants of the TP53 gene that are not strongly linked to cancer.
- The rs1042522 (Arg72Pro) SNP creates two TP53 gene variants / alleles - 'C' (Pro) and 'G' (Arg).
- The different 'C' (Pro) and 'G' (Arg) variants are shown to alter p53 activity and affect relative effectiveness of DNA repair, cell cycle arrest and apoptosis.
How do common TP53 variants affect p53 function?
As mentioned earlier, there are three key ways by which p53 responds to DNA damage and prevents damaged cells from replicating:
- DNA repair – activation of enzymes that fix various forms of DNA damage
- Cell cycle arrest – preventing the cell from progressing through the cell cycle to disallow cell division.
- Apoptosis – elimination of cells with heavy DNA damage through programmed cell death.
The relative activity of these three processes may be affected by what TP53 gene variants we inherit.
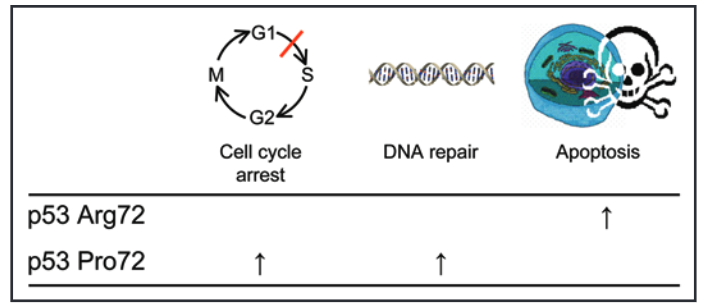
Source: Bojesen, S. E., & Nordestgaard, B. G. (2008). The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. cell cycle, 7(2), 158-163.
On this note, studies have shown that the ‘G’ or Arg72 allele of the TP53 gene codes for a p53 protein that is more effective at promoting apoptosis, but less efficient at DNA repair and cell cycle arrest.
By contrast, the ‘C’ or Pro72 allele encodes a p53 protein that is relatively more effective at DNA repair and cell cycle arrest.
Interestingly, this ‘C’ / Pro72 allele has been linked to greater lifespan and survival after life-threatening illness, an effect which may be due to better DNA repair and cell cycle arrest. Furthermore, while apoptosis is undoubtedly crucial for destroying damaged and potentially cancerous cells, it has been speculated that less readily-activated apoptosis could be beneficial, as it allows cells to regenerate after some forms of tissue injury (e.g. ischaemic damage) rather than be eliminated.
KEY POINTS
- The 'C' (Pro) allele of the TP53 gene codes for a p53 protein that is relatively more effective at DNA repair and cell cycle arrest.
- The 'G' (Arg) allele codes for a p53 protein that is relatively better at activating apoptosis - programmed cell death.
- TP53 gene variants may have an effect on lifespan and longevity.
How do TP53 variants affect lifespan and longevity?
An analysis of 9,219 Danish subjects enrolled in the Copenhagen City Heart study found that those carrying the ‘C’ or Pro allele had a longer lifespan and reduced mortality after a cancer diagnosis.
In the study, subjects had blood samples to assess their TP53 genotype and then were followed up for 12 years. Researchers then looked at who survived across this 12-year period and analysed whether survival corresponded to differences in TP53 genotypes.
As illustrated by the shallower gradients of the green and red lines in the left-hand graph below, a much greater proportion of those with the Pro/Pro (CC) and Arg/Pro (GC) genotypes survived compared to those with the Arg/Arg (GG) genotype.
More specifically, those with the Pro/Pro (CC) and Arg/Pro (GC) genotypes had a 6% and 3% higher 12-year survival than the Arg/Arg (GG) genotype, respectively. When adjusting for sex and age, this translated to a 18% (CC) and 12% (GC) lower risk of death from any cause compared to the GG (Arg/Arg) genotype. Doing more number crunching, this corresponded to 3 and 2 years greater median survival for CC (Pro/Pro) and GC (Arg/Pro) genotypes.
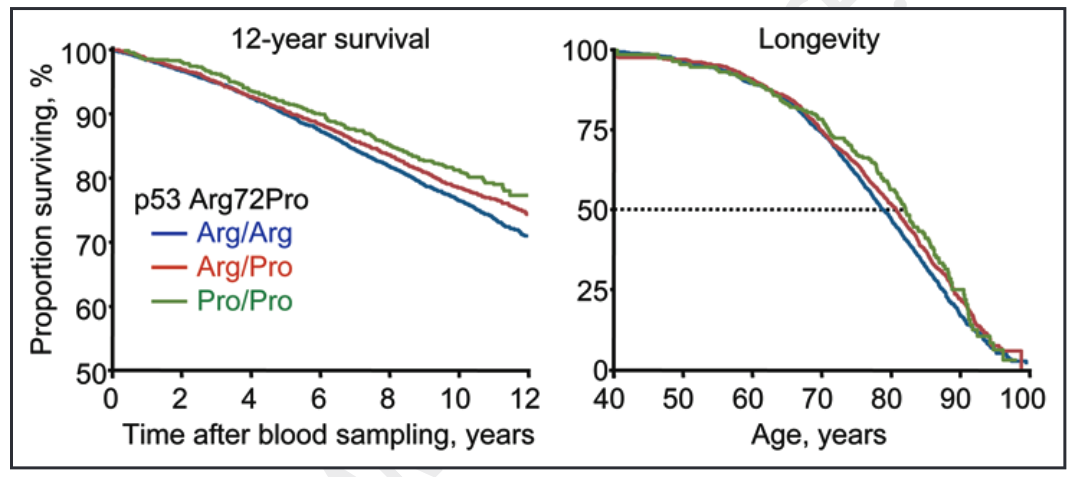
Source: Bojesen, S. E., & Nordestgaard, B. G. (2008). The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. cell cycle, 7(2), 158-163.
So, why did ‘C’ (Pro) allele carriers have a longer lifespan?
Interestingly, the researchers did not find any significant association between TP53 genotype and risk of developing cancer, cardiovascular disease, or other life-threatening illness.
Rather, the study found that, after developing cancer or other life-threatening illness, ‘C’ (Pro) allele carriers had better survival. On this note, those with the CC (Pro/Pro) and GC (Arg/Pro) genotypes were found, respectively, to have a 13% and 9% lower cumulative 5-year mortality following a cancer diagnosis.
This beneficial effect of the ‘C’ / Pro allele on survival can be seen in the graph below, with a greater proportion of CC (Pro/Pro) (green line) and GC (Arg/Pro) (red line) surviving in the 5 years following a cancer diagnosis.
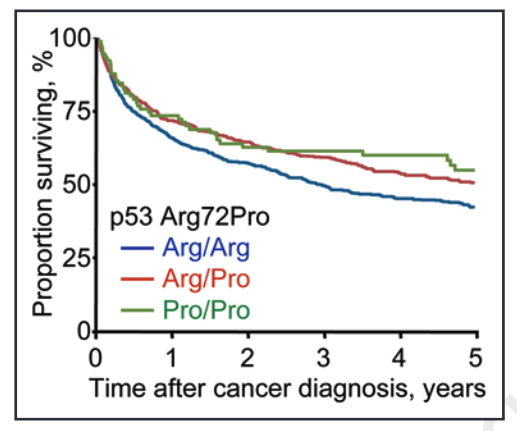
Source: Bojesen, S. E., & Nordestgaard, B. G. (2008). The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. cell cycle, 7(2), 158-163.
The study also found ‘C’ (Pro) allele carriers to have greater survival following Type II diabetes, lung disease, and infections. Another analysis, focussing on elderly subjects in the Leiden 85-plus Study, found that those with the CC (Pro/Pro) genotype had a 41% increased survival rate, with greater proportions of those with this genotype surviving past 85 years old.
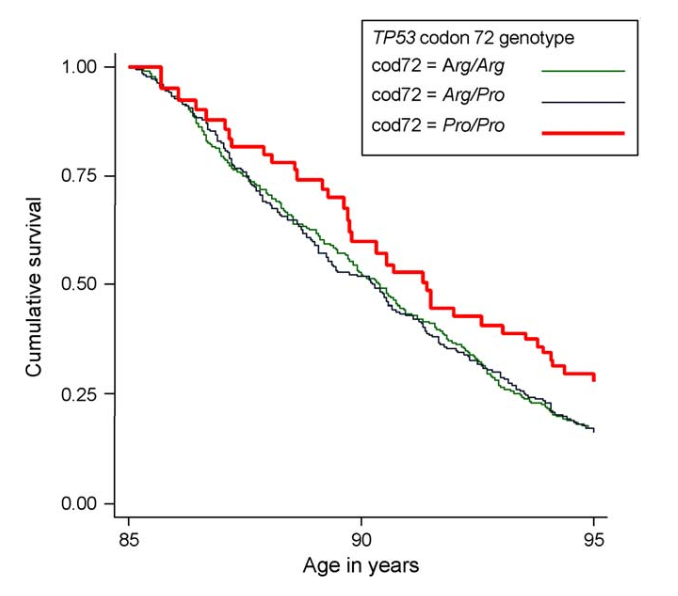
Source: van Heemst, D., Mooijaart, S. P., Beekman, M., Schreuder, J., de Craen, A. J., Brandt, B. W., ... & Westendorp, R. G. (2005). Variation in the human TP53 gene affects old age survival and cancer mortality. Experimental gerontology, 40(1-2), 11-15.
The reasons behind this relationship between the ‘C’ (Pro) allele of the TP53 gene and longevity are unclear and further research is required into the underlying biological mechanisms.
As the ‘C’ allele codes for a p53 protein that is more efficient at activating DNA repair and cell cycle arrest, it is possible that this affords greater resilience to life-threatening illness, such as cancer. Similarly, as previously alluded to, it is also possible that reduced activation of apoptosis in ‘C’ / Pro allele carriers may also enhance survival of cells following damage.
KEY POINTS
- Some studies have linked the 'C' (Pro) allele of the TP53 gene to greater lifespan and longevity.
- The 'C' (Pro) allele has also been linked to greater survival after a cancer diagnosis.
- These effects may be due relatively more effective DNA repair and cell cycle arrest in 'C' (Pro) allele carriers.
- Greater activation of apoptosis in 'G' (Arg) allele carriers may impair regeneration of cells following some types of damage, although this is not proven.
How do common TP53 variants affect cancer risk?
As the p53 protein plays a key role in protection against uncontrolled cell growth and the formation of tumours, we might expect common TP53 gene variants to affect cancer risk.
Generally speaking, for a cell to become cancerous and divide uncontrollably, the following must happen:
- It must accumulate DNA damage that is unrepaired.
- It must escape arrest at cell cycle checkpoints.
- It must evade apoptosis.
Based on this, we might expect the ‘C’ (Pro) allele, which is linked to enhanced DNA repair and cell cycle arrest, to confer greater protection against cancer compared to the ‘G’ (Arg) allele.
On the contrary, it also makes sense that the ‘G’ (Arg) allele, which may more effectively activate apoptosis of heavily damaged precancerous cells, confers greater protection against cancer.
Studies of various cancers so far paint a mixed picture. Some studies have found an association between the ‘G’ (Arg) allele and a higher risk of cancer. For example, a meta-analysis of studies in Sub-Saharan populations found the ‘G’ (Arg) allele was associated with a higher risk of cervical cancer.
By contrast, other studies have linked the ‘C’ (Pro) allele to a greater cancer risk. For example, a 2020 meta-analysis found a greater risk of breast cancer in carriers of the ‘C’ (Pro) allele.
Several other studies have found no link between TP53 genotype and cancer risk. A meta-analysis of studies looking at melanoma, a type of skin cancer, did not find any significant difference in risk when comparing TP53 genotypes.
Again, it is important to reiterate here that the common TP53 variants based on the rs1042522 (Arg72Pro) SNP are not robustly linked to cancer. Even in the studies reporting significant associations between TP53 genotype and cancer risk, the odds ratios reported are fairly small, suggesting only mildly increased susceptibility.
We can contrast this with the rare inherited mutations of the TP53 gene, not analysed at FitnessGenes, that can give rise to Li-Fraumeni syndrome. These rare mutations cause defective p53 function, leading to much greater susceptibility to cancer. In fact, men and women with Li-Fraumeni syndrome have a ≥70% and ≥90% risk of cancer, respectively, in their lifetimes.
KEY POINTS
- Studies linking common TP53 gene variants to cancer risk are mixed.
- Some studies have suggested the 'G' (Arg) allele confers greater protection against various cancers.
- Other studies suggest the opposite, the 'C' (Pro) allele is linked to lower cancer risk.
- Several studies report no association between TP53 genotype and cancer risk.
- The effects of the 'G' (Arg) and 'C' (Pro) alleles - both common TP53 gene variants - on cancer risk are likely to be small.
- Rare TP53 mutations can give rise to Li-Fraumeni Syndrome, which is characterised by very high risk of developing cancer.
- FitnessGenes do not analyse mutations linked to Li-Fraumeni syndrome.
Your DNA repair and longevity trait
Your DNA repair and longevity trait looks at the rs1042522 (Arg72Pro) SNP in the TP53 gene. Based on your DNA results, you will be classified into one of three groups:
- Reduced DNA repair: you carry two copies of the ‘G’ (Arg) allele linked to less effective DNA repair and cell cycle arrest, but more readily activated apoptosis. Your genotype (GG) is not linked to increased longevity.
- Moderately increased DNA repair: you carry one copy of the ‘C’ (Pro) allele linked to more effective DNA repair and cell cycle arrest, and greater longevity.
- Increased DNA repair: you carry two copies of the ‘C’ (Pro) allele linked to more effective DNA repair and cell cycle arrest, and greater longevity. This genotype (CC) group is linked to the greatest longevity.
To find out your result, please login to truefeed.




.png)
